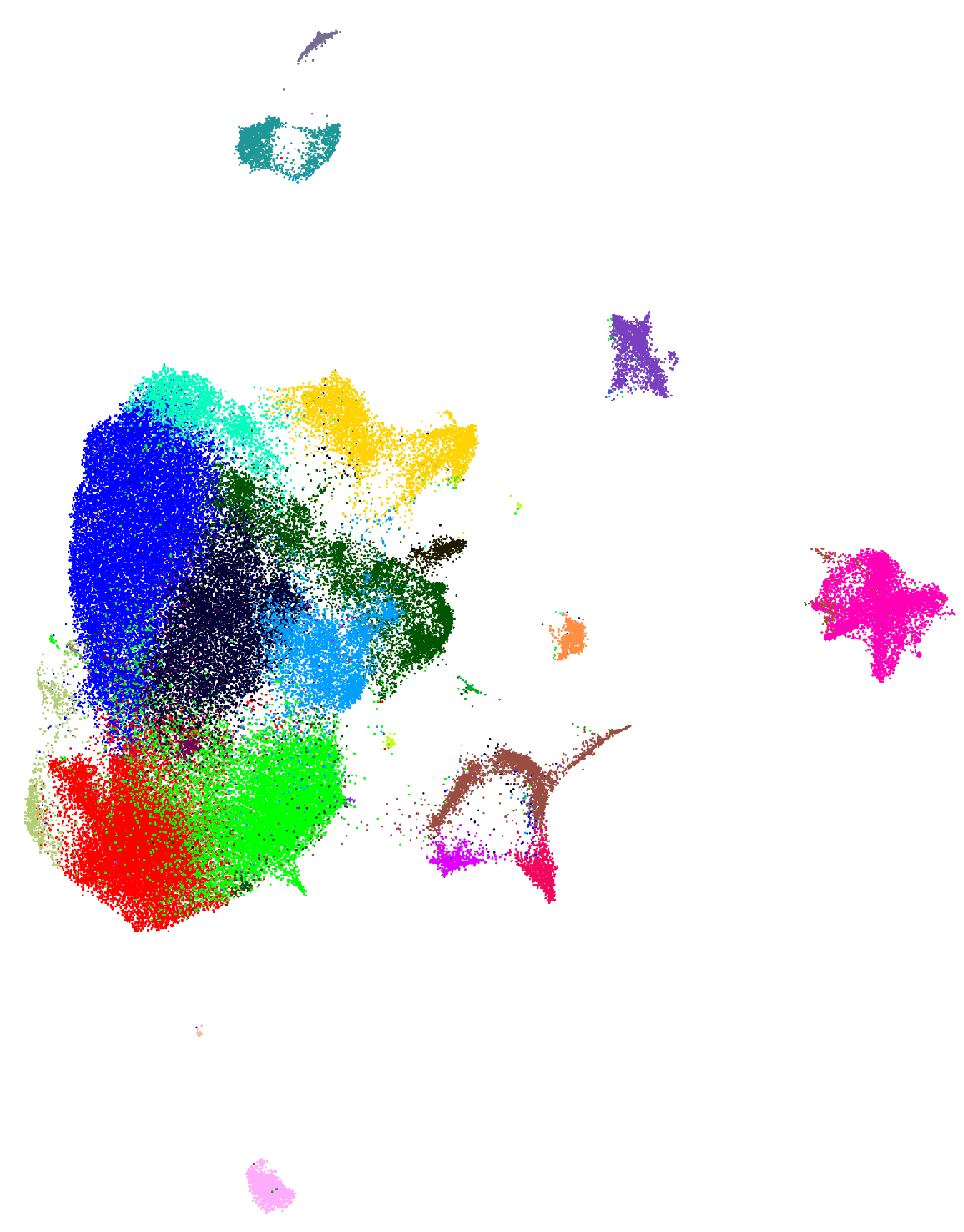1.How do pathogens cause plant disease? I investigate the functionalities of fungal effectors using fungal genetics and plant pathology to elucidate how pathogens adapt to host defenses and manipulate plant physiology. Through transcriptomics and epigenetics, I define the transcriptional patterns of effectors during infection and dissect the regulatory mechanisms that precisely control their expression. My recent work discovered that a transcriptional regulator suppresses a group of effectors in M. oryzae, governing the crosslink between appressoria development and transcriptional regulation during invasive growth.
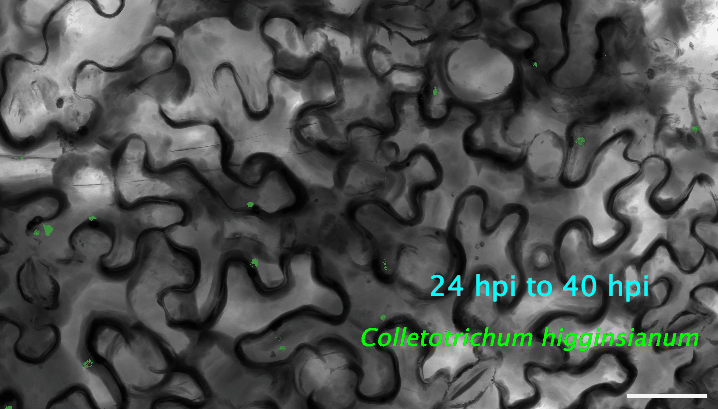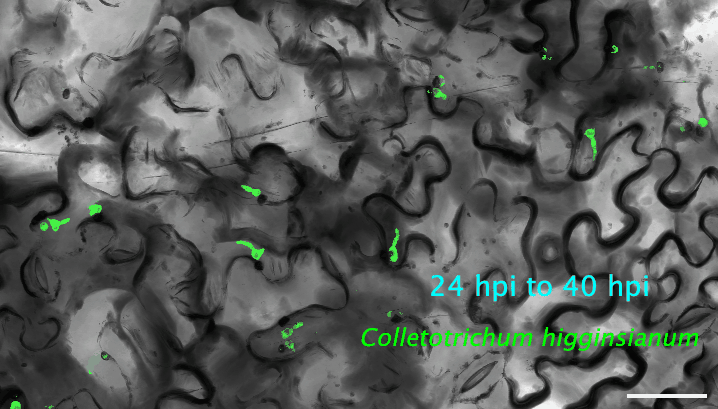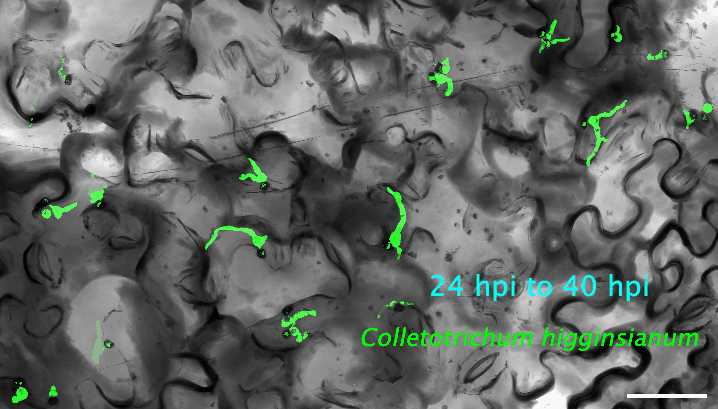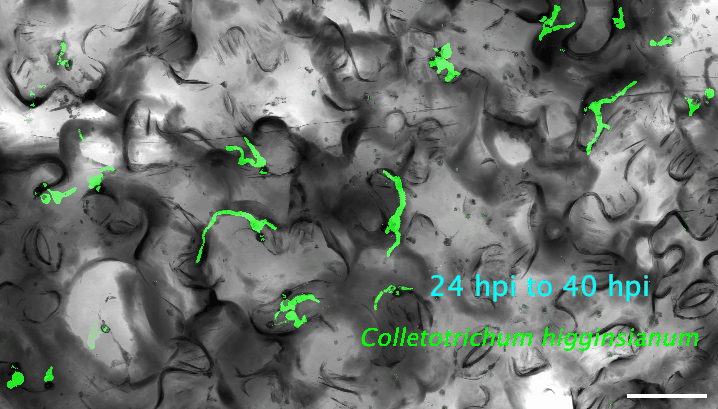
2. How do plant cells respond to infection at the single-cell level? I use cutting-edge approaches, such as single-cell transcriptomics and AI-based imaging, to study the molecular dynamics and heterogeneity in different plant cell types in response to pathogen infection. I determine the precise deployment of immune receptors in various plant tissues and cell types. Additionally, I reconstruct the spatiotemporal dynamics of plant-fungus interactions, enabling the identification of infection site cells that directly contact fungal invasive hyphae. This facilitates uncovering high-resolution molecular dynamics and metabolic signatures at the interfaces between plants and pathogens. My recent work revealed a fine-tuned deployment of immune receptors in the plant vasculature system. The next step of my research is to determine the regulatory elements of cell-type-specific expression of immune receptors. The ultimate goal is to mine and/or deploy crop cultivars with robust resilience against various stresses without growth penalties or fitness costs.